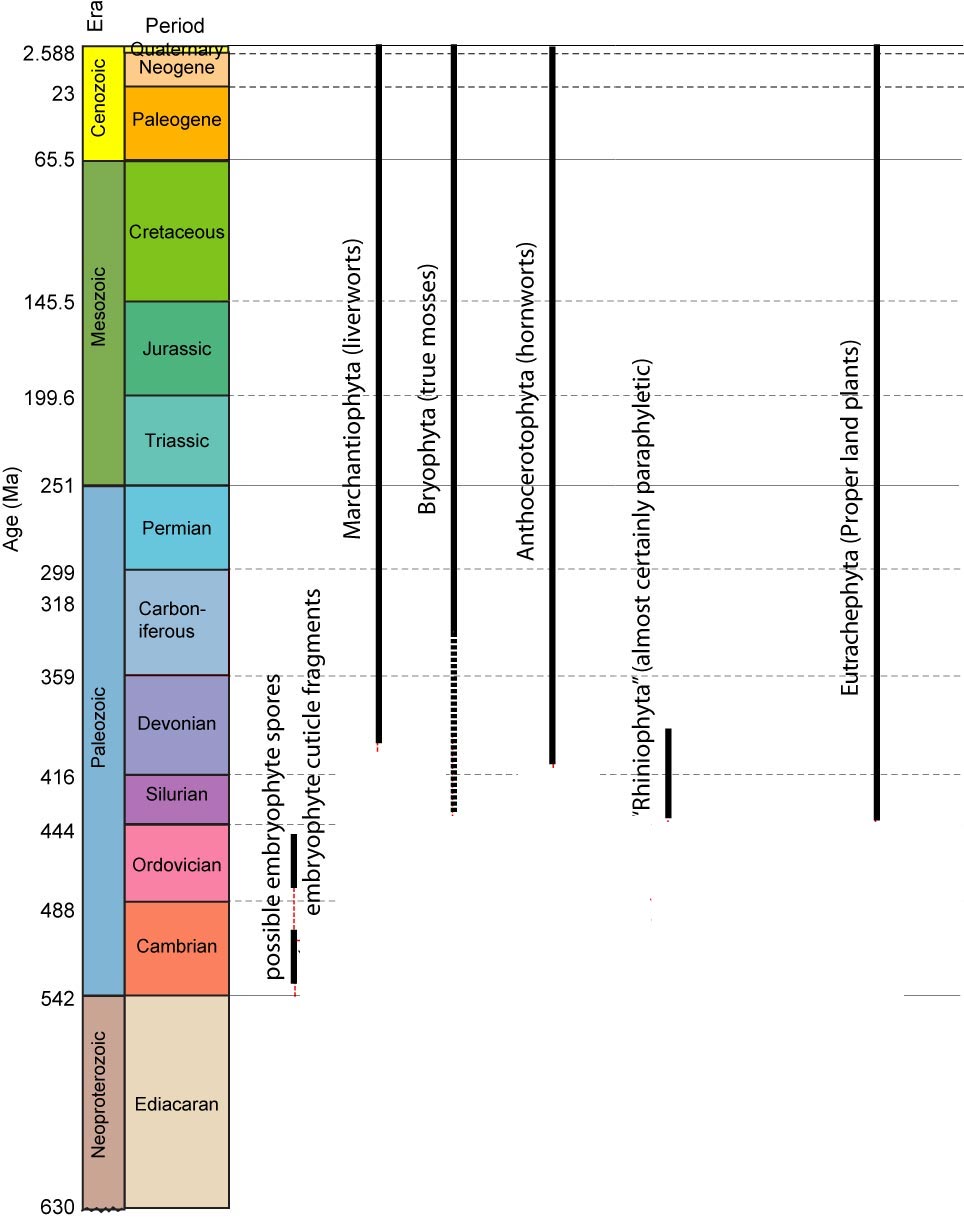
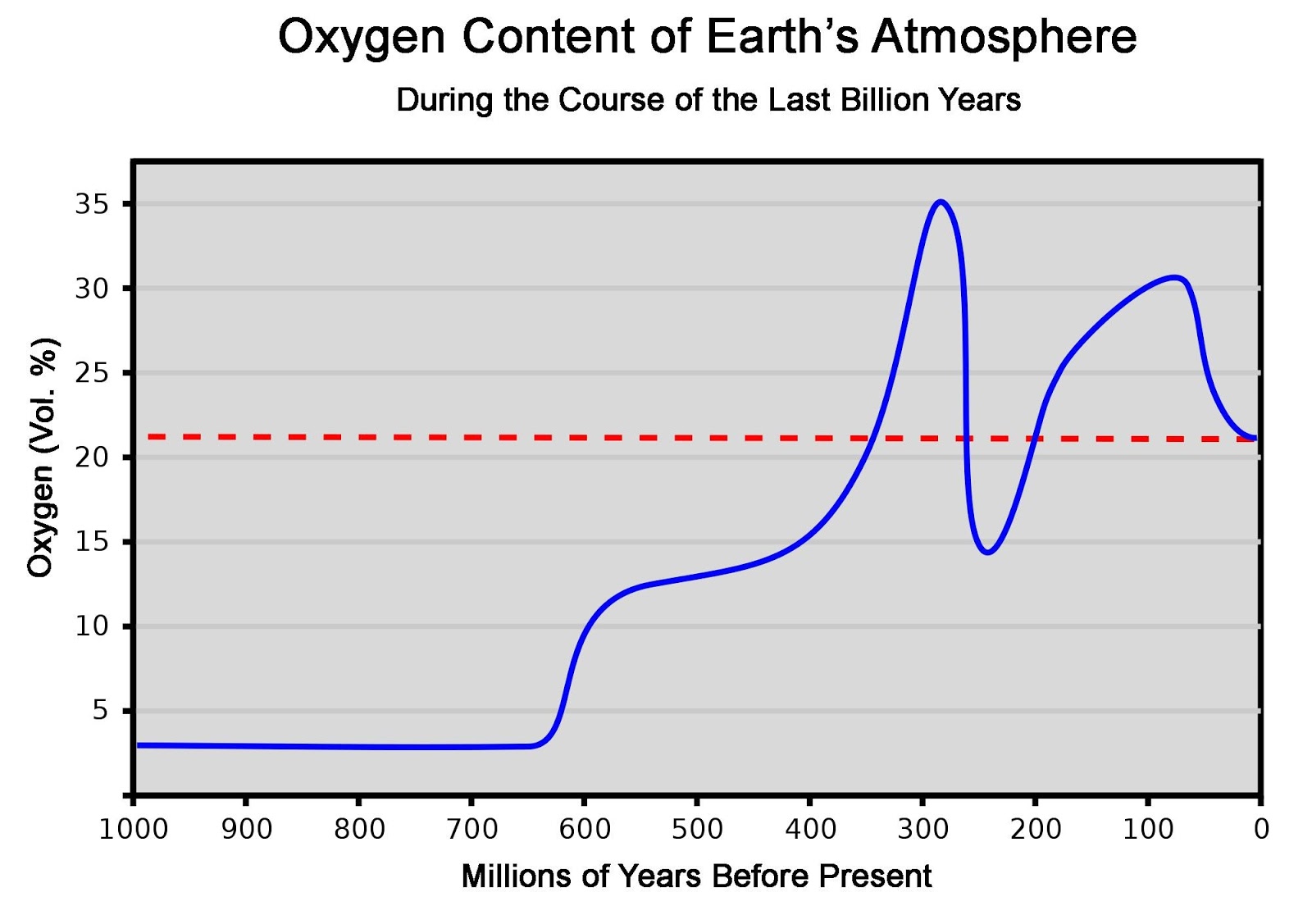
Sunday, November 24, 2013
Wednesday, October 9, 2013
Wednesday, July 31, 2013
Monday, June 17, 2013
Sunday, May 26, 2013
Monday, April 15, 2013
Saturday, March 23, 2013
Thursday, March 21, 2013
Tuesday, February 26, 2013
Gorilla Genome EYA4 Marker vs. Evolution
is the EYA4 Marker.
In general, deletions are more likely than deletions.
However you slice it, the genetic information contradicts current constructions.
Saturday, February 23, 2013
Human - Chimp Chromosome Comparisons
Both articles claiming 96% similarity between Humans and Chimps
are from 2005, i.e., from seven years ago.
This is widely known to be out-of-date data.
Since the first crude attempt to sequence the chimp genome on a budget
less than 1/10 that used for the human genome project,
it has come to light that the methodology was faulty and contamination
was evident.
Genome size data for chimps and humans using flow cytometry
of cell nuclei to estimate nuclear DNA content, indicates that the chimp genome
may be up to 10% larger than the human genome (www.genomesize.com)
The most recent version of the chimp genome assembly at www.ensembl.org
confirms the data from flow cytometry studies showing that
the chimp genome is AT LEAST 8% larger than the human genome.
These are the real up-to-date data on genome-size.
The article I referenced, which IS up-to-date in regard to at least one
whole chromosome, freshly sequenced, was just published in 2010.
The new 2010 article shows an addition 1.5% divergence specifically in the Y-chromosome section.
96% - 1.5% = 94.5%.
More disinformation.
But the other links clearly show an additional 8% of the Chimp Genome was also neglected
in the total genome comparison, because it was simply discarded in that and the discussion.
94.5% - 8% = 86.5%
More disinformation.
But the earlier discussion of how the original 98% estimates were guesstimated
shows that less than 5% of the actual genome was compared in those estimates.
So those estimates were worthless in the first place.
More disinformation discovered.
The final 2006 comparisons with the shoddily constructed chimp genome (fuller version)
were also unreliable, because the chimp genome was reconstructed on a doubly
faulty basis, first using the Human Genome as the map instead of doing a chimp map,
and second because of contamination of the chimp samples.
More disinformation discovered.
While we are listening to Evolutionists claim that the
entire Human / Chimp genome has been allegedly compared in detail (as of 2005),
we should note carefully
what the 2010 article reports in passing
on the topic of complete Genome comparisons:
16. Watanabe, H. et al. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429, 382–388 (2004) | Article |
They conveniently provide the map, which indeed shows the strong similarity in chromosome 21 (one arm only!):
Here the strong diagonal line on the right (chromosome 21 long half)
indeed shows how strongly the Human and Chimp DNA aligns
(The dots however are not contiguous, and greater resolution would show holes).
Now look at the Y-Chromosome comparison. NOTHING lines up in position,
there IS no orientation that can give maximum alignment,
and half the content has no mapping at all between Human and Chimp.
This is NOT a simple case of some empty repeats 'tacked on the end',
as some have tried to misleadingly claim.
Most of the non-alignable regions contain both protein-coding genes
(7 families not found in chimps) and bizarre anagram-code (on the chimp side)
which is indeed functional, although only just discovered and hardly understood.
What the data shows plainly, is that humans are indeed from planet earth,
but that the chimp Y-chromosome may be from Mars.
Conspiracy theorists and Alien Visitor investigators will be all over this,
for the next 20 years. Because there is nothing like it in the genome
of ANY animals so far sequenced.
The other projection from the investigative article shows even more clearly
the very suspicious recombination cleavage artifacts,
which indicate that this chromosome was artificially cut in fixed lengths
having nothing to do with the content of the chromosome, or its functional
sections. The authors may have been too disturbed by its implications
or potential misinterpretations to put it in the main article.
The data strongly suggests that the chimp is an artificially gene-customized lab animal.
Such constructs obviously won't conveniently fit on any evolutionary tree at all.
There is another explanation however, that is the most plausible, but will be the least popular:
that the databases for the chimp have been compromised by shoddy low budget methodology,
which makes them useless for comparisons where the Chimp Genome has indeed been obliterated.
but what is in fact far more likely,
is that the 'matches' to the human genome are false positives
due to contamination of the chimp database with human DNA.
entire Human / Chimp genome has been allegedly compared in detail (as of 2005),
we should note carefully
what the 2010 article reports in passing
on the topic of complete Genome comparisons:
"...chromosome 21, [is] the only other chromosome comprehensively mapped and sequenced in both species16. "
16. Watanabe, H. et al. DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429, 382–388 (2004) | Article |
They conveniently provide the map, which indeed shows the strong similarity in chromosome 21 (one arm only!):
Quote:
| Each dot represents 100% chimpanzee–human identity within a 200-base-pair (bp) window. In the Y-chromosome plot, the human chromosome is oriented with short arm to top and long arm to bottom, and the chimpanzee chromosome is oriented with short arm to left and long arm to right. For chromosome 21, which is acrocentric, the plot represents only the long arm. |
Here the strong diagonal line on the right (chromosome 21 long half)
indeed shows how strongly the Human and Chimp DNA aligns
(The dots however are not contiguous, and greater resolution would show holes).
Now look at the Y-Chromosome comparison. NOTHING lines up in position,
there IS no orientation that can give maximum alignment,
and half the content has no mapping at all between Human and Chimp.
This is NOT a simple case of some empty repeats 'tacked on the end',
as some have tried to misleadingly claim.
Most of the non-alignable regions contain both protein-coding genes
(7 families not found in chimps) and bizarre anagram-code (on the chimp side)
which is indeed functional, although only just discovered and hardly understood.
What the data shows plainly, is that humans are indeed from planet earth,
but that the chimp Y-chromosome may be from Mars.
Conspiracy theorists and Alien Visitor investigators will be all over this,
for the next 20 years. Because there is nothing like it in the genome
of ANY animals so far sequenced.
The other projection from the investigative article shows even more clearly
the very suspicious recombination cleavage artifacts,
which indicate that this chromosome was artificially cut in fixed lengths
having nothing to do with the content of the chromosome, or its functional
sections. The authors may have been too disturbed by its implications
or potential misinterpretations to put it in the main article.
The data strongly suggests that the chimp is an artificially gene-customized lab animal.
Such constructs obviously won't conveniently fit on any evolutionary tree at all.
There is another explanation however, that is the most plausible, but will be the least popular:
that the databases for the chimp have been compromised by shoddy low budget methodology,
which makes them useless for comparisons where the Chimp Genome has indeed been obliterated.
but what is in fact far more likely,
is that the 'matches' to the human genome are false positives
due to contamination of the chimp database with human DNA.
Thursday, February 21, 2013
Evolution: Wrong
Lets briefly go over the incredible LOGIC FAILS
that Evolutionists like gcthomas succumb to:
(1) On the Non-Existence of Self-Replicating Molecules:
Rather than refute the plain argument and evidence against
the possibility of an INANIMATE object as small as a 200 Nucleotide string
being able to 'magically' reproduce itself from a non-biotic environment,
gcthomas quotes biologists actually using the phrases "replicating",
and "self-replication", along with anthropomorphic language that
attributes motivation to dead chemical objects.
He completely fails to grasp that this is evidence of fraudulent presentation
and unscientific 'magical thinking',
not any kind of evidence for the existence of a 'self-replicating molecule'.

(2) On Human and Chimp DNA being allegedly 98% identical
An actual biologist defending evolution quotes another evolutionist-biologist,
admitting that the figures were way off, and that Human and chimp DNA
are NOT even close to 98% identical.
Another geneticist of 15 years experience explains in detail why
Human and Chimp DNA can't be closer than 70% similar.
gcthomas responds that Human and Chimp DNA are actually then 96% identical.
Are we seeing a pattern of inability to comprehend
plain statements, and learn new things?

that Evolutionists like gcthomas succumb to:
(1) On the Non-Existence of Self-Replicating Molecules:
Rather than refute the plain argument and evidence against
the possibility of an INANIMATE object as small as a 200 Nucleotide string
being able to 'magically' reproduce itself from a non-biotic environment,
gcthomas quotes biologists actually using the phrases "replicating",
and "self-replication", along with anthropomorphic language that
attributes motivation to dead chemical objects.
He completely fails to grasp that this is evidence of fraudulent presentation
and unscientific 'magical thinking',
not any kind of evidence for the existence of a 'self-replicating molecule'.

(2) On Human and Chimp DNA being allegedly 98% identical
An actual biologist defending evolution quotes another evolutionist-biologist,
admitting that the figures were way off, and that Human and chimp DNA
are NOT even close to 98% identical.
Another geneticist of 15 years experience explains in detail why
Human and Chimp DNA can't be closer than 70% similar.
gcthomas responds that Human and Chimp DNA are actually then 96% identical.
Are we seeing a pattern of inability to comprehend
plain statements, and learn new things?

I just received a copy in the mail today of Dr. Thomas' new book,
"More Than A Monkey: The Human - Chimp DNA Similarity Myth" (2012).
Here's an excerpt from the Introduction:
"More Than A Monkey: The Human - Chimp DNA Similarity Myth" (2012).
Here's an excerpt from the Introduction:
Quote:
"While I was aware that very little support existed in the DNA sequence data
for Darwinian macro-evolution, the supposed nearly identical DNA similarity
between human and chimpanzee as touted by the popular press,
many scientists, and even some professing Christian scientific authorities,
was obviously an issue that needed more scrutiny and research.
The task at first seemed daunting, but the deeper I delved into studying
the published scientific literature and even performing my own genomic analyses,
the more I realised that the human-chimp DNA similarity data had been grossly filtered,
manipulated, cherry-picked and obfuscated to meet an academic
gold standard of political correctness originally
set during the 'early days' of bio-technology.
In fact, ... a history of the human-chimp 'nearly identical' DNA paradigm is covered in chapter 1."
Monday, February 18, 2013
Dolphins and Cows Again
Now lets examine the implications of the model, and the data:
12 differences / 100 bases = The cow differs from its common ancestor with the dolphin in 12% of its DNA bases.
7 / 100 = The dolphin only differs from their common ancestor in 7% of DNA bases.
Since it is agreed that each species mutated in quite different directions,
and that backtracking is virtually impossible under normal conditions,
its a safe bet that:
virtually all DNA mutations in the Dolphin line that move it
away from the common ancestor also move it away from the cow.
Few if any of those mutations will 'reverse' DNA evolution or bring
the species closer together. Even the sites of mutation will be different,
virtually guaranteeing that the mutations will all be unique, and non overlapping.
The measure of genetic 'distance' from the central (common ancestor) point,
can simply be added together, or even more plainly, placed on a horizontal
'distance' numberline:
Now however, the connection point to the rest of the phylogenetic tree
must be decided. This is done on the basis of a crude, literal 'parsimony',
taking the DNA alterations at face value.
As a scientific procedure however, this has extremely dubious value,
because there is no 'one-to-one' correspondence between a nucleotide change,
and a real feature or characteristic of an actual species or individual.
Many mutations have no effect at all, and no informational value,
and no relevance to Speciation in particular or Evolution in general.
Such mutations give 'Natural Selection' nothing to work with,
and the only measurable effect is a slow, relatively benign 'genetic drift'.
A REAL attempt at a meaningful and probable location-point for common
ancestry and speciation would take into account both the morphology and
the glaring differences in function between animals, which would
interact with the environment to actually drive Natural Selection in
a plausible and comprehensible fashion.
It is trivially obvious that a cow is far more similar to a horse, dog, cat, and alpaca, than it is to a dolphin, and the crude and literal measure of
'DNA differences' at a nucleotide level is both misleading and useless.
Environmental changes drive Natural Selection, and we can expect that
drastic environmental changes force rapid Natural Selection, i.e.,
punctuated equilibrium style effects, and such is the very example before us.
The following phylogenetic connection takes into account rationally
both what the theory of Evolution predicts, and what the data really
indicates. If the mutations themselves are semi-random processes,
only constrained by survival and reproductive forces, then they will
likely target critical and non-critical segments of DNA in the same proportions
that these appear.
Since it is Natural Selection itself which determines which DNA modifications
are adopted generationally, its obvious that there should be more DNA changes
where Natural Selection can be expected to be more intensive and active.
This tree places the 'common ancestor' at a point which will account for
the expected differences in mutation rates, caused by the different intensities
of Natural Selection operating in a background milieu of Genetic Drift.
It further poses the likelihood that the common ancestor of the cow and dolphin
was a land animal, unremarkable, and much closer to its other near relatives,
such as horse, alpaca, dog and cat, than to sea creatures.
The difference between the two reconstructions is simply that
the genetic data is not blindly applied in a clumsy and mindless fashion,
but is interpreted by the Theory of Evolution itself, and integrated into
an intelligent picture of the speciation process.
Although this is not any kind of evidence in favour of the Theory of Evolution,
it is necessary to impose upon any interpretation of data (genetic or otherwise),
the constraints which the Theory itself poses.
12 differences / 100 bases = The cow differs from its common ancestor with the dolphin in 12% of its DNA bases.
7 / 100 = The dolphin only differs from their common ancestor in 7% of DNA bases.
Since it is agreed that each species mutated in quite different directions,
and that backtracking is virtually impossible under normal conditions,
its a safe bet that:
virtually all DNA mutations in the Dolphin line that move it
away from the common ancestor also move it away from the cow.
Few if any of those mutations will 'reverse' DNA evolution or bring
the species closer together. Even the sites of mutation will be different,
virtually guaranteeing that the mutations will all be unique, and non overlapping.
The measure of genetic 'distance' from the central (common ancestor) point,
can simply be added together, or even more plainly, placed on a horizontal
'distance' numberline:
Now however, the connection point to the rest of the phylogenetic tree
must be decided. This is done on the basis of a crude, literal 'parsimony',
taking the DNA alterations at face value.
As a scientific procedure however, this has extremely dubious value,
because there is no 'one-to-one' correspondence between a nucleotide change,
and a real feature or characteristic of an actual species or individual.
Many mutations have no effect at all, and no informational value,
and no relevance to Speciation in particular or Evolution in general.
Such mutations give 'Natural Selection' nothing to work with,
and the only measurable effect is a slow, relatively benign 'genetic drift'.
A REAL attempt at a meaningful and probable location-point for common
ancestry and speciation would take into account both the morphology and
the glaring differences in function between animals, which would
interact with the environment to actually drive Natural Selection in
a plausible and comprehensible fashion.
It is trivially obvious that a cow is far more similar to a horse, dog, cat, and alpaca, than it is to a dolphin, and the crude and literal measure of
'DNA differences' at a nucleotide level is both misleading and useless.
Environmental changes drive Natural Selection, and we can expect that
drastic environmental changes force rapid Natural Selection, i.e.,
punctuated equilibrium style effects, and such is the very example before us.
The following phylogenetic connection takes into account rationally
both what the theory of Evolution predicts, and what the data really
indicates. If the mutations themselves are semi-random processes,
only constrained by survival and reproductive forces, then they will
likely target critical and non-critical segments of DNA in the same proportions
that these appear.
Since it is Natural Selection itself which determines which DNA modifications
are adopted generationally, its obvious that there should be more DNA changes
where Natural Selection can be expected to be more intensive and active.
This tree places the 'common ancestor' at a point which will account for
the expected differences in mutation rates, caused by the different intensities
of Natural Selection operating in a background milieu of Genetic Drift.
It further poses the likelihood that the common ancestor of the cow and dolphin
was a land animal, unremarkable, and much closer to its other near relatives,
such as horse, alpaca, dog and cat, than to sea creatures.
The difference between the two reconstructions is simply that
the genetic data is not blindly applied in a clumsy and mindless fashion,
but is interpreted by the Theory of Evolution itself, and integrated into
an intelligent picture of the speciation process.
Although this is not any kind of evidence in favour of the Theory of Evolution,
it is necessary to impose upon any interpretation of data (genetic or otherwise),
the constraints which the Theory itself poses.
Sunday, February 17, 2013
Killing Evolution with DNA (2)
We are now in a position to understand why phylogenetic trees
never propose LIVING species as direct descendants of one another.
The DNA evidence is damning for any such real construction,
because the Genome of EVERY species shows plain 'cross-mixture',
unique to almost every triplet of species chosen.
All ancestors must be hypothetical any systems which attempt
to give credibility to the hypothesis of No DNA Transfer between species.
All ancestors and nodes must be hypothetical in nature,
and based on key assumptions like the impossibility of inter-species DNA transfer.
But this also means that all primary DNA-based phylogenetic trees
can only be one level deep. Further backward constructions must
continue building upon already hypothetical constructions, with
less and less reliability and certainty.
Such hypothetical phylogenetic trees must make connections based
on a kind of 'mutation Parsimony' which attempts to minimize the
number of mutations. This itself is the best policy, but it cannot be
assumed that genetic mutations have always travelled the simplest path.
But the distances portrayed on these reconstructed 'trees'
have another glaring fault. They assume that mutation rates are
more or less constant, predictable, and can be used as reliable indicators
of 'evolutionary distance' and even estimated time.
Thus a typical phylogenetic tree will indicate the number of mutations
using a distance scale, inferring also relative timing of evolutionary steps
of hypothetical speciation.
Such a tree is very misleading however, because the Theory of Evolution itself demands that real preserved mutations be transferred at widely varying rates,
according to the environmental factors driving Natural Selection.
In reality, these phylogenetic trees constructed using DNA information
have been artificially extended and made to look more 3-dimensional,
when in actual fact they are quite flat and unable to comment on the actual historical rates of transmitted DNA mutations and timings of speciation events.
A less misleading presentation of the same data would look like this:
Here we see what the DNA evidence is really saying:
(1) The tree is only one generation deep as far as living organisms,
because real organisms cannot be placed in a credible descent tree.
(2) The number of mutations counted along hypothetical routes,
back to hypothetical 'nodes' of hypothetical ancestors has no precise
meaning in terms of time or even mutation rates.
This is because according to the theory, only Natural Selection
can determine which mutations become passed on and what the
timing will be.
never propose LIVING species as direct descendants of one another.
The DNA evidence is damning for any such real construction,
because the Genome of EVERY species shows plain 'cross-mixture',
unique to almost every triplet of species chosen.
All ancestors must be hypothetical any systems which attempt
to give credibility to the hypothesis of No DNA Transfer between species.
All ancestors and nodes must be hypothetical in nature,
and based on key assumptions like the impossibility of inter-species DNA transfer.
But this also means that all primary DNA-based phylogenetic trees
can only be one level deep. Further backward constructions must
continue building upon already hypothetical constructions, with
less and less reliability and certainty.
Such hypothetical phylogenetic trees must make connections based
on a kind of 'mutation Parsimony' which attempts to minimize the
number of mutations. This itself is the best policy, but it cannot be
assumed that genetic mutations have always travelled the simplest path.
But the distances portrayed on these reconstructed 'trees'
have another glaring fault. They assume that mutation rates are
more or less constant, predictable, and can be used as reliable indicators
of 'evolutionary distance' and even estimated time.
Thus a typical phylogenetic tree will indicate the number of mutations
using a distance scale, inferring also relative timing of evolutionary steps
of hypothetical speciation.
Such a tree is very misleading however, because the Theory of Evolution itself demands that real preserved mutations be transferred at widely varying rates,
according to the environmental factors driving Natural Selection.
In reality, these phylogenetic trees constructed using DNA information
have been artificially extended and made to look more 3-dimensional,
when in actual fact they are quite flat and unable to comment on the actual historical rates of transmitted DNA mutations and timings of speciation events.
A less misleading presentation of the same data would look like this:
Here we see what the DNA evidence is really saying:
(1) The tree is only one generation deep as far as living organisms,
because real organisms cannot be placed in a credible descent tree.
(2) The number of mutations counted along hypothetical routes,
back to hypothetical 'nodes' of hypothetical ancestors has no precise
meaning in terms of time or even mutation rates.
This is because according to the theory, only Natural Selection
can determine which mutations become passed on and what the
timing will be.
How to Kill Evolution using DNA
One necessary requirement for proving a specific line of Common Descent
among species is the presumed impossibility of 'mixture' across species.
Without this constraint, DNA could then hop across populatons
from species to species freely, and any gene or piece of code
could come from anywhere. Tracing DNA would be impossible.
To test whether DNA could hop between species is actually very simple.
For our purposes, we'll call this the Transitivity Test. If three species fail the Transitivity Test, no specific line of Common Descent is possible,
and some other explanation must be posited for the results of the test.
An example will make this clear:
We examine a common strip of DNA shared by three species.
Much of the strip is both identical and shared by all three,
so we are reasonably sure we are looking at the same zone:
(1) (2) (3) (4)
__________________________________________________________
Species A: .. CT.--- TGGA --- CAT --- CTGAGCCCC...
Species B: ...GT--- TGGA --- CGT --- CTGAGCCCC...
Species C: ...GT--- CGGG --- CAT --- CTGAGCCCC...
__________________________________________________________
Suppose we want to test a specific possible Descent Tree:
We propose that Species C is the common ancestor of A and B:
The problem is, each species shares some DNA with the others:
But according to our primary axiom,
that Species cannot exchange DNA directly,
this is already an impossible situation.
In fact, it is impossible, no matter which species
we make the 'common ancestor'.
One might say 'no problem; these three species
are not direct descendants of one another.
But unfortunately, it does not end here:
Not only is it impossible for these species to be directly descended,
it also requires that we actually explain how they came to share DNA,
if they were NOT descended from one another.
There can only be two basic possibilities:
(1) They all inherited their DNA from a single common ancestor,
and the unique shared 'readings' indicate mutation (corruption of DNA), or,
(2) By coincidence two species happened to have their DNA
mutated or corrupted at the very same spot and in exactly the same way!
Neither of these cases bode well for genetic methods of tracing genealogy:
If (1) is allowed, then no tree can be constructed with any certainty
in regard to descent, since we already can see that the DNA data is
too corrupted to use to construct genealogies.
The frequency or commonness of a reading is no longer any indication
of "when" it originated or even "if" a given reading is original or a corruption.
If (2) is allowed, we have to recognize something even worse;
We might explain a single corruption of one Nucleotide (one letter)
by mere bad luck: Once a mistake occurs in the same spot,
the mistake has a 1 in 4 chance of matching a mistake in the other species. But to match in exactly the same place is a whole order of magnitude more unlikely. In real cases, we are going to find thousands of such 'mistakes',
stretching for much longer sequences than a single Letter in the DNA code.
These kinds of 'coincidences' become so unlikely as to make them
practically impossible.
In any case, it is preferable to uphold the hypothesis of a barrier
preventing DNA transfer between species generally, since it is
an observable trend, whereas coincidental mutations are statistically
highly implausible.
But the cost is twofold:
(1) no direct tree of descent will be allowable.
(2) A hypothetical 'common ancestor' (now lost) must always be added.
But this is not any kind of evidence that a common ancestor even existed.
Instead, many other possibilities are still left wide open, such as common building blocks, mechanical constraints, or a shared design.

With the less likely option, i.e., (2) Cross-species DNA exchange,
we have to contrive a new mechanism instead of a new ancestor.
In order to account for significant correspondences between two species,
not shared by other species, we can postulate another mechanism,
that would allow the transmission of DNA between species, defeating
speciation and its premise entirely.
The cost here is as follows:
(1) The acceptance of a hypothetical transfer mechanism between species.
(2) The abandonment of the axiom that DNA is not transferred between species.
In practice it is impossible to determine which case explains any given
set of matches. Even if it were, the very fact of "mixture" makes
the reconstruction of specific genealogical trees of descent impossible.
But if NO case of genetic inheritance can be traced to descent,
then there can be no evidence of Evolution and descent from DNA analysis.
Saturday, February 16, 2013
REAL Evidence for Evolution
Alate_One and other geniuses here don't understand exactly what would constitute
good evidence for common descent.
What all the examples given have in common is their incompleteness,
which is actually fatal to any claim of their being evidence for common descent.
To see why this is, we can take a simple parallel case
with hand-copied manuscripts.

If manuscripts were simply copied, and never compared and corrected,
then tracing the genealogy of any manuscript would be child's play.
Each time an 'error' or variation is introduced into the copying transmission line,
it would be copied by subsequent scribes and all descendents of that manuscript
from that point on would contain the error.
Even if a small number of copies in the lines were lost,
we could still reconstruct the original archetype or common ancestor
with near-certainty.
In fact, even if we only had the very last generation of copies,
and all earlier ones were destroyed, we could reconstruct with good reliability
the original text, as long as certain assumptions were made about
the number of copies made from each manuscript, and just how
representative of the whole transmission line (for the entire period)
that the final sample (the last generation) of manuscripts was.
The assumption would be made that manuscripts on average
were copied about the same number of times,
and also that the error rate (accidental changes introduced into the copying line)
was also more or less constant.
On the basis of those two assumptions,
we could look at variations, and make deductions about the original text.
For instance, if only 10 manuscripts had one reading,
and 90 manuscripts had an alternate reading at some place,
we could assume that the minority reading was a late error,
which only affected 10% of the copying stream.
On this basis, we could create a hypothetical copying tree,
and even 'date' the errors, or at least put them in a temporal order,
based upon how strong the reading was (what % of copies contained it).

_________________________________________________
The genetic information in DNA is a similar case:
(1) The DNA is copied and transmitted generation to generation.
(2) Occasional errors are introduced.
(3) Subsequent copies contain the errors.
(4) The 'error rate' is presumed to be reasonably constant.
(5) Errors cannot be corrected once introduced, because the DNA can only be genetically inherited.
______________________________________
So much for theory, now lets look at practice,
based on the real situation on the ground:
In the case of manuscript copying,
(1) Copies are also proofread and corrected before release into the population.
(2) Copies are often corrected not just from their own master-copy,
but also from earlier copies, and even copies from a different line of transmission.
(3) Cross-pollination of readings makes tracing genealogies virtually impossible,
even if we were to possess every copy or source in every
transmission line. It is impossible for instance, to know which copy
a scribe used in correcting the text, when thousands of manuscripts
already would have had the reading used.

(4) This is the phenomenon called in Textual Criticism "mixture".
This contamination process not only makes it impossible to construct
the real genealogies of manuscripts, but it also makes it impossible
to even accurately group or classify manuscripts, because of their 'mixed'
groups of readings, which are often randomly scattered in the texts.
Sometimes clusters of manuscripts seem well-behaved,
sometimes they simply don't:

__________________________________________
Now lets turn to the problem of DNA genealogies:
Inside the Same Species:
(1) Within a species, sexual reproduction in higher species means that
mixture of DNA lines is constantly taking place, and this mixture makes
genealogy impossible to trace beyond a few generations. It also makes
it impossible to strictly classify individuals in a population according to
their DNA coding.
(2) The very limited additional constraints imposed by Mitochondrial transmission
and Y-chromosome transmission (through sexual reproduction)
for higher animals, combined with the short time-spans,
make genealogical tracing very limited and of little practical value.
Currently geneticists can only make general statements extending back
100,000 years or so.
(3) Furthermore, Recombination further mixes DNA transmission lines,
making genealogical tracing impossible.
(4) Finally however, the genealogical relationship between members of the
same species is not in doubt, and neither is it any kind of evidence for
the Theory of Evolution, which is intended to explain the appearance of
different species.
Between Species:
(1) Outside a species, mixture CANNOT in theory take place, since
it is impossible for DNA lines to mix once again, after speciation
has occurred and cross-breeding is no longer a viable channel of mixture.
(2) However, genealogical relationships between completely different species
are the very thing to be proved. Evidence must be independently
procured for genealogical dependence and descent between species,
before DNA data can be reliably applied.
(3) Like the case with ancient manuscripts, typically we only have access to
the DNA of currently living animals or recently deceased species,
i.e., the last few generations of DNA copying.
(4) This means that all genealogical relationships between species
must be reconstructed working backward from current DNA samples
of living species, if DNA evidence is to be used.
(5) In order for temporal ordering of DNA mutations to be accomplished,
the rates of change must be assumed to be constant, or non-volatile,
i.e., changing slowly and reliably.
(6) At the same time, in order to construct real time-lines, the mutation rates
must not only be known to be constant, but they also must be known
in absolute terms, so that time periods can be estimated, and distant
parallel branches of DNA mutation can be ordered.
(7) However, Natural Selection as a process must operate in response to
real changes in the environment and so the ultimate transmission of DNA
mutations CANNOT be constant, but must be responsive to actual ground conditions
and so actual TRANSMITTED mutations must be volatile and unpredictable in reality.
(8) Furthermore, the current evidence of punctuated equilibrium is
not a mere artifact of an impoverished fossil record, but is a very real
phenomenon predicted by the Theory of Evolution. If accepted,
this fundamental fact negates not only the assumption of fixed DNA
mutation rates, but prevents any hierarchical ordering of specific changes
on a real timeline, either in parallel branches, or consequently in direct
lines of descent.
(9) External principles of 'parsimony' or other methodology must be used
in order to organize the information of DNA differences themselves.
Hence, DNA evidence cannot be used to directly support any theory
of genealogical relationship, nor any specific reconstructions of the
relationships between species.
________________________________________
If DNA evidence can't prove Evolution, i.e., speciation, descent,
or even a temporal order of events, let alone a real timeline,
nor can it independently corroborate other independent evidence
of descent, what can it do?
Remarkably, Genetic information provided by DNA actually
can do something very important:
Genetic information can disprove or falsify specific claims made
by the Theory of Evolution.
How?
Simple:
If DNA evidence demonstrated probable evidence of "mixture",
when speciation in fact prohibits mixture,
then the Theory of Evolution is dead in the water:
Some other explanation for genetic similarities,
sharing of DNA information, would have to be proposed.
In order for genetic information to disprove Evolution,
complete comparisons are required,
between more than two species:
(1) Genetic information could be shared between any two species.
(2) Identical genetic information could be shared between three or more species.
(3) However, DIFFERING genetic information could NOT be shared
between two overlapping pairs of species, because it would be impossible
to explain how the 'mixture' occurred.
(4) This is the 'acid test' for Evolutionary theory, because the theory offers
no mechanism whatever for how significant DNA information could cross
between any two species AFTER speciation has occurred between the three different species.
____________________________________________
Lets see why this would utterly disprove Evolution;
Take the parallel case of manuscript copying:
If two late copies shared the same reading, there is only one of three
possibilities to explain it;
(1) Either it is the original reading, shared by common descent,
(2) Mixture has occurred, and a reading was borrowed from another line,
(3) Its a remarkable coincidence: two scribes independently made the same mistake.
Since (2) Mixture is impossible between species, only (1) and (3)
are viable options to explain the phenomenon.
But as the number of potential cases of 'mixture' increase,
the likelihood of 'accidental coincidence' (i.e., identical mutations)
drops dramatically, and in fact quickly to the point of impossible.
The alternate explanation left behind, is that
some other cause for the shared DNA sequences must be postulated,
namely physical design constraints, or mechanical borrowing,
or re-use of identical parts in assembly.
________________________________
This is the reason why human / chimp DNA sequences are
not reviewed in total, but in small samples, like partial samples
of mitochondrial DNA, or singe genes.
This is also the reason why Evolutionists avoid comparing
more than one species at a time in total.
The obvious 'mixture' phenomena would be apparent to all,
and the Theory of Evolution would be dead in the water.
Gene Fusion and Mitochondria
Even the Field of Genetics however was annoyingly ambiguous about Man's descent from early apes.
The one exciting piece of evidence that seemed to relate Man to primates generally,
was Man's chromosome count, which seemed to indicate an early fusion
of two genes.
But unfortunately, this evidence was worthless in addressing the
key issue in the question of the descent of man from apes:
When and how did it happen?
It turns out that the supposed 'fusion' must have taken place
when 'Man' was already 'evolved' into a Man, some 5 million years
AFTER the alleged 'mutation/evolution' of Man from early Hominid ancestors.
Worse than that, this 'fusion' apparently had no significant effect on
the actual change of 'Man' into something else.
Its not an actual mutation of genes per se, but
simply a reorganization of exactly the same DNA content,
and might only affect the ability of the recipient to mate with
other humans. (a mere sterility issue).
So it wasn't germane to the main event after all.
All it definitively proved, was that all men (excepting mutations)
were indeed descended from the same ancestor with 23 chromosome pairs.
Mitochondrial Con-Jobs
Evolutionists next turned to the new 'Mitochondrial' evidence;
but unfortunately that simply didn't help either, because it
merely bracketed the problem with brackets FAR TOO WIDE
to have anything meaningful to say on Descent of Man:

(1) On the one hand, (according to the latest evolutionary theory)
living things were supposed to have taken on mitochondria in an event
in the very very distant past, so that all modern living things larger
than a single cell contain them. (whether even this theory has merit is
another question).
(2) On the other hand, tracing the mitochondrial mutations only seems
to allow confident genealogical connections going back a few hundred
thousand years, nowhere near the needed 10 million years to when
supposedly men evolved from apes.
Another dud-cannon missing the mark, by firing over both bow and stern.
The one exciting piece of evidence that seemed to relate Man to primates generally,
was Man's chromosome count, which seemed to indicate an early fusion
of two genes.
But unfortunately, this evidence was worthless in addressing the
key issue in the question of the descent of man from apes:
When and how did it happen?
It turns out that the supposed 'fusion' must have taken place
when 'Man' was already 'evolved' into a Man, some 5 million years
AFTER the alleged 'mutation/evolution' of Man from early Hominid ancestors.
Worse than that, this 'fusion' apparently had no significant effect on
the actual change of 'Man' into something else.
Its not an actual mutation of genes per se, but
simply a reorganization of exactly the same DNA content,
and might only affect the ability of the recipient to mate with
other humans. (a mere sterility issue).
So it wasn't germane to the main event after all.
All it definitively proved, was that all men (excepting mutations)
were indeed descended from the same ancestor with 23 chromosome pairs.
Mitochondrial Con-Jobs
Evolutionists next turned to the new 'Mitochondrial' evidence;
but unfortunately that simply didn't help either, because it
merely bracketed the problem with brackets FAR TOO WIDE
to have anything meaningful to say on Descent of Man:

(1) On the one hand, (according to the latest evolutionary theory)
living things were supposed to have taken on mitochondria in an event
in the very very distant past, so that all modern living things larger
than a single cell contain them. (whether even this theory has merit is
another question).
(2) On the other hand, tracing the mitochondrial mutations only seems
to allow confident genealogical connections going back a few hundred
thousand years, nowhere near the needed 10 million years to when
supposedly men evolved from apes.
Another dud-cannon missing the mark, by firing over both bow and stern.
Phylogenetic Trees again
The first real "Knight in Shining Armor" to come and rescue the 'Princess Evolution"
was the new 'science of tree making', comprised of three different but
competing, (and inevitably contradictory) fields:

When these three stooges failed to rescue the princess,
along came the exciting new field of genetics.
was the new 'science of tree making', comprised of three different but
competing, (and inevitably contradictory) fields:
"There are three currently active schools of taxonomy:(cf. Conceptual Issues in Evolutionary Biology, - Sokol, pg 239 f)
(1) phenetic taxonomy,
(2) cladistics
(3) evolutionary systematics."

When these three stooges failed to rescue the princess,
along came the exciting new field of genetics.
Descent Trees of Man
Not put off by getting their hands caught red-handed in the till of making shiite up,
the Evolutionists promptly turned instead back to 'Trees of Descent'.
Of course here once again, the biased projections began to stand out like
a sore thumb against the actual fossil evidences and gaping holes for
the needed 'transitionals':
As the early hominid evidence multiplied,
the picture just became more and more 'complicated',
rather than the expected 'filling in of the blanks'.
That is because the evidence simply didn't fit the Evolutionist picture
being packaged and sold.
It was like having to ram more and more square pegs into round holes:
Even the 'Evolutionary Trees of Descent' started to become
more and more disjointed and disconnected,
leaving the public in a confused state:
Subscribe to:
Comments (Atom)